Presented By: Ecology and Evolutionary Biology
EEB Virtual Seminar: Mapping biochemical drivers of phenotypic change
Dr. Daniel Jarosz, Associate Professor of Chemical and Systems Biology and of Developmental Biology, Stanford University
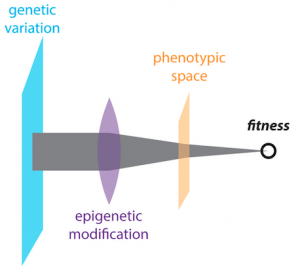
Survival in changing environments requires the acquisition of new heritable traits. However, mechanisms that safeguard the fidelity of DNA replication often limit raw material available to fuel such novelty. Our research centers on an intriguing resolution to this paradox: intrinsic links between the folding of biological polymers and virtually every trait in an organism provide multiple avenues through which environmental stress can impact evolution, disease, and development. Stress-regulated molecular chaperones of protein folding such as Hsp90, can broadly influence whether genetic variants can produce new phenotypes. We have identified other proteins, including a protein chaperone of RNA folding, that act in a conceptually similar fashion, ‘buffering’ the consequences of accumulating mutations. Other work focuses on the 30% of eukaryotic proteins that do not adopt a single fixed structure. Many mutations associated with metazoan innovation and human diseases occur within such intrinsically disordered proteins (IDPs). Yet the function of this ‘dark matter’ of the proteome is largely unknown. We recently found that IDPs involved in gene control can commonly adopt self-templating conformations – molecular ‘memories’ heritable over long biological timescales. This echoes the paradigm-shifting biology of prion proteins. But the ~50 we discovered frequently drive adaptive gains-of-function and do not form amyloid. Most remarkably, their self-templating capacity is often conserved from yeast to humans, leading us to propose that protein-based ‘molecular memories’ might commonly drive new interpretations of the genetic code. In addition to illuminating a new form of epigenetics ‘beyond the chromosome,’ understanding the difference between physiological and pathological protein aggregation has important implications for many intractable human pathologies.
Join us on Zoom
Join us on Zoom